The console at the bottom of the YGOB interface allows a user to select:

- Which genomes to display by selecting and unselecting them.
- What version of YGOB to use.
- What window size to display. This varies from ±4 genes up to ±50. The larger the window size the longer the browser will take to display the information.
- Which gene to focus the display on.
- Whether to turn RNA features on or off (from Version 4).
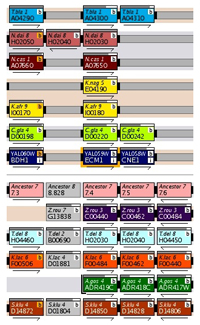 Gene Name Box
Gene Name BoxIn YGOB genes are represented by boxes featuring gene and chromosome information. Each genome has a colour pallette (red, green, blue etc.) which is used to distinguish genes from different chromosomes or contigs - genes without synteny are coloured grey.
From Version 4 additional features are displayed on screen as well. Centromeres are displayed by black boxes, RNA features (e.g. tRNA genes) are shown by white boxes and Ty proteins are coloured dark grey.
In-Focus Gene
The gene in focus is highlighted by a orange border. A user can click on any other gene to refocus YGOB on it.
Arrows
The arrows under each gene box denote the gene's relative transcriptional orientation.
'Length' Bars (from Version 7)
In Version 7 we introduced bars at the top of gene boxes that shows the relative length of homologs, normally coloured grey, they are coloured salmon if the gene contains an intron. Rolling over the bar gives the exact percentage and length.
Species and Strain Names (from Version 8)
These are displayed in a sidebar to the right of each track.
Connectors join nearby genes - a solid connector for adjacent genes, two small bars for genes within 5 genes of each other and one small bar if they are within 20 genes. These connectors are usually coloured black, but are highlighted in orange if they denote an inversion. The connectors are continued over any intervening space between genes with grey extensions.
From Version 3 of YGOB clicking on solid connectors or their grey extensions (i.e. connectors between adjacent genes in a genome) links to the intergenic sequence between those two genes.
Brackets
The end of a chromosome or contig is denoted by a bracket around the gene's box.
Banding
Alternating grey and salmon bands behind the tracks in YGOB link genomes from the same evolutionary clade.
In Version 3 of YGOB we introduced a small number of genes we have annotated ourselves in YGOB. These are all of the form '[Genome Short Code]_YGOB_[Gene Name]' and so are easily recognised. In Versions 4 & 5 we annotated hundreds of such genes in a large-scale search for unannotated genes.
From Version 3 of YGOB we introduced the ability of curators to turn off annotated but dubious features, these however remain in the annotation files (flagged with 'OFF') in case users are interested in them. In addition, clicking through to an intergenic region containing such a feature will display a note of its existence.
 Each gene box has a "b" button which when clicked opens a window with BLAST (blastp) results for that gene against the yeast genomes in the browser. Results are highlighted in red if they are in the same column as the gene, blue if they appear on
screen outside of its column, and orange if they are a tandem copy of the gene.
Normally coloured white, the "b" button is orange when the gene has a tandem repeat on screen nearby.
Each gene box has a "b" button which when clicked opens a window with BLAST (blastp) results for that gene against the yeast genomes in the browser. Results are highlighted in red if they are in the same column as the gene, blue if they appear on
screen outside of its column, and orange if they are a tandem copy of the gene.
Normally coloured white, the "b" button is orange when the gene has a tandem repeat on screen nearby.
In more recent versions of YGOB we have added three extra columns to the output, one showing the ON/OFF status of the hit, one showing an abbreviation of the species the gene is from, and another showing the hit's ancestral pillar, if any. Hits from genomes not selected for display are listed in italic. Users can also now select some or all of the listed hits for processing by YGOB's various bioinformatics tools.
Extracts the protein sequences for genes in a column.
"nt" Button (from Version 3)
Extracts the nucleotide sequences for genes in a column.
"msa" Button (from Version 3)
 Outputs a MUSCLE multiple sequence alignment of the the protein sequences of the genes in a column.
Outputs a MUSCLE multiple sequence alignment of the the protein sequences of the genes in a column.
"tree" Button (from Version 3)
Draws a phylogenetic tree for the genes in a column using a MUSCLE alignment and PhyML.
"rates" Button (from Version 3)
Generates pairwise yn00 output of Ka, Ks and omega for all genes in a column.
With the ever growing number of genomes in YGOB most are not displayed by default. A number is parathesis by the species/strain name to the right of each track indicates the total number of genomes from that genus that are available for display, "(2)", "(8)" etc. Clicking on the number will expand the genus and add tracks for each genome from it. When a genus is fully expanded a "(-)" button is displayed - clicking it collapses the genus to just the track you clicked beside (if the focused gene is from another genome in that genus it will remain too).
S. cerevisiae gene boxes have an "i" information button than clicks through to the SGD title line description of the protein and its Gene Ontology terms. Rolling your mouse over a gene's box will also display any available information for that gene.
Every visualised YGOB page has a "+" button in its bottom left hand corner. Clicking the button displays the same YGOB data in a tabulated text format.
The control console has a "Flip" button to allow the user to swap the top and bottom tracks.
"Twist" Button
The control console has a "Twist" button to allow the user to invert the left-right sequence of the columns in the display.
Each column has a "S" button at its base, which clicks through to the protein sequences for the genes in that column.
"T" Button (Versions 1 & 2)
Any column containing three or more genes has a "T" button at its top, which draws an approximate phylogenetic tree for the genes in that column using a T-Coffee alignment and Phylip's neighbour joining program.
Some of these may only work in later versions of YGOB.
As well as understanding systematic names for S. cerevisiae genes (e.g. YAL063C), YGOB will also correctly identify SGD genetic names (e.g. FLO9 in the case of YAL063C) if inputted, as well as synonyms. In the former case it will bring you straight to the gene, in the latter case you will be asked to confirm the gene you are interested in. YGOB will also take as input the 'stem' of a SGD systematic name (e.g. YAL064) and present the list of genes with that stem (YAL064W-B, YAL064C-A, YAL064W in this example).
In many genomes it may not be obvious but the choosen naming convention what are the first or (especially) the last genes on a chromosome. Typing in the genome's four letter reference code (lowercase with leading letter in uppercase), followed by an underscore, followed by the chromosome number, then a period and then 'L' or 'R' will take you to the first or last gene on that chromosome, respectively.
To get to a given co-ordinate position on a chromsome for a given genome, type in the genome's four letter reference code (uppercase), followed by a colon, followed by the chromosome number, then another colon, and then the kilobase position of interest (i.e. 100 means base 100,000). The browser will bring you to the gene at or adjacent to that co-ordinate.
If you know the number of a pillar of interest (they are used mainly as internal IDs in YGOB but can be seen embedded in various links) and wish to navigate to it (without having to recall a gene in the pillar) then type in 'P' followed by a pipe character ('|') followed by the pillar number. YGOB will bring you to the pillar with the browser focused on a gene in it (a non-WGD gene if one is present). This is mainly a tool for curators. Pillar numbers may not remain the same from version to version.
 The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species
The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid speciesByrne KP and Wolfe KH
Genome Research. 2005 Oct;15(10):1456-61
| Supplemental: | Methods | Table 1 | Table 2 |
 Visualizing syntenic relationships among the hemiascomycetes with the Yeast Gene Order Browser
Visualizing syntenic relationships among the hemiascomycetes with the Yeast Gene Order BrowserByrne KP and Wolfe KH
Nucleic Acids Research. 2006 Jan 1;34(Database issue):D452-5.
